Biography
I am a Scientific Python Developer working at Zuru Tech.
I previously worked as a postdoctoral researcher in the Group of Roberto Cerbino at the Faculty of Physics of the University of Vienna, and in the group of Francesco Sciortino at the Physics Department of the University of Rome La Sapienza. My academic research interests include the investigation of the structure and dynamics of complex colloidal suspensions using advanced optical techniques and the development of high-performance codes for experiments analysis (using Python and C++/CUDA).
- High-performance computing with Python, C++, and CUDA
- Structure, dynamics, self-assembly, and phase-separation in complex fluids
- Advanced optical techniques applied to soft matter
PhD in Industrial Chemistry and Chemical Engineering, 2019
Polytechnic of Milan
MSc in Nuclear Engineering, 2015
Polytechnic of Milan
BSc in Energy Engineering, 2012
Polytechnic of Milan
Experience
- Experimental investigation and analysis of non-equilibrium effects in sedimentation
- Development of fastDDM to accelerate differential dynamic microscopy analysis
- Laboratory establishment
- Development of optical setup for the characterization of the dynamics of colloidal samples
- Development and characterization of DNA gels for biomedical applications
- Molecular dynamics simulations of DNA nanoparticles
Skills
Projects
Recent Posts
Featured Publications
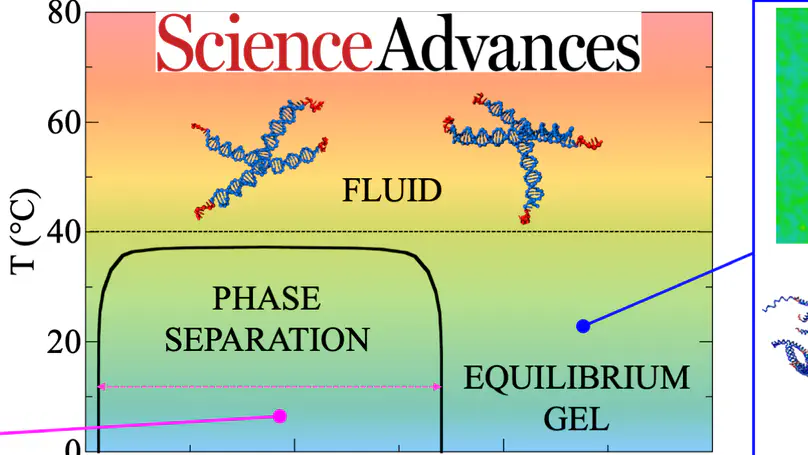
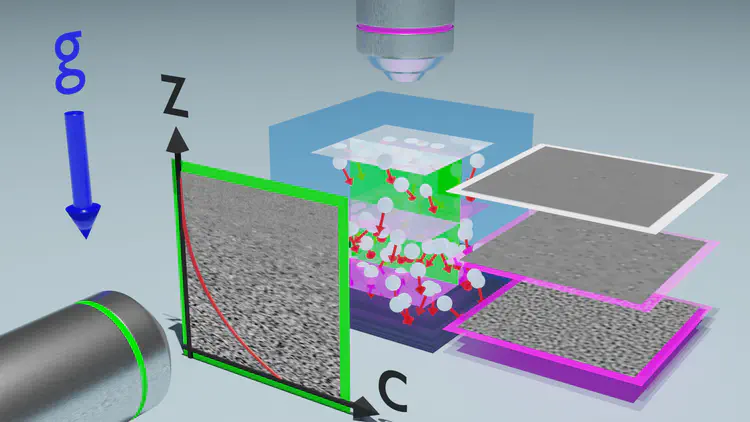
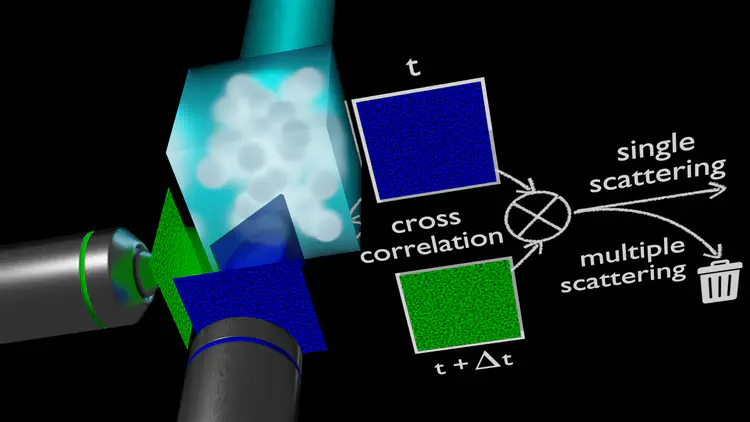
Gels of DNA nanostars, besides providing a compatible scaffold for biomedical applications, are ideal model systems for testing the physics of equilibrium colloidal gels. Here, using dynamic light scattering and photon correlation imaging (a recent technique that, by blending light scattering and imaging, provides space-resolved quantification of the dynamics), we follow the process of gel formation over 10 orders of magnitude in time in a model system of tetravalent DNA nanostars in solution, a realization of limited-valence colloids. Such a system, depending on the nanostar concentration, can form either equilibrium or phase separation gels. In stark contrast to the heterogeneity of concentration and dynamics displayed by the phase separation gel, the equilibrium gel shows absence of aging and a remarkable spatially uniform dynamics.
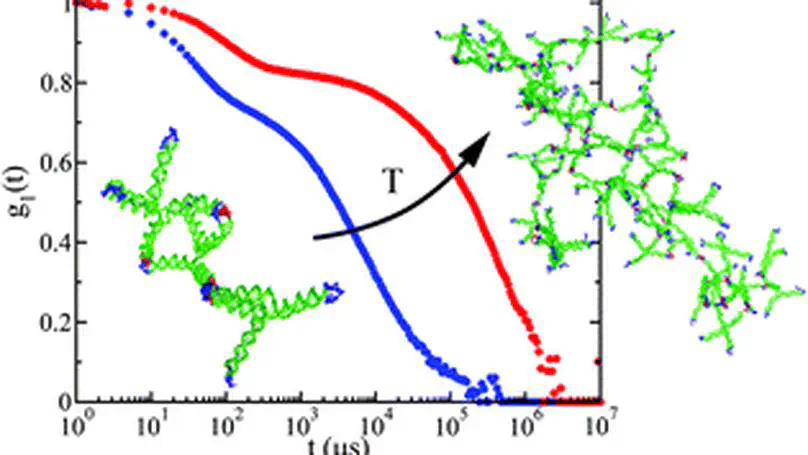
Taking advantage of the base-pairing specificity and tunability of DNA interactions, we investigate the spontaneous formation of hyperbranched clusters starting from purposely designed DNA tetravalent nanostar monomers, encoding in their four sticky ends the desired binding rules. Specifically, we combine molecular dynamics simulations and Dynamic Light Scattering experiments to follow the aggregation process of DNA nanostars at different concentrations and temperatures. At odds with the Flory–Stockmayer predictions, we find that, even when all possible bonds are formed, the system does not reach percolation due to the presence of intracluster bonds. We present an extension of the Flory–Stockmayer theory that properly describes the numerical and experimental results.
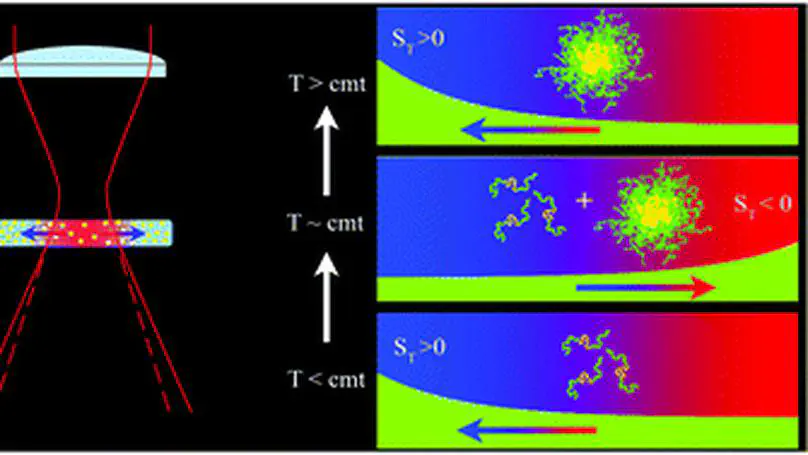
Due to its exquisite sensitivity to interfacial properties, thermophoresis, i.e., particle motion driven by thermal gradients, can provide novel, exclusive, and often surprising information on the structural properties of colloidal or macromolecular fluids and on particle/solvent interactions at the nanoscale. Here, by using an all-optical thermal excitation technique, thermal lensing, we show that thermophoresis can be profitably exploited to investigate the self-association of an amphiphilic block copolymer, poloxamer P407, which takes place above a concentration-dependent critical micellization temperature (cmt). In particular we show that, around and above the cmt, the direction of the poloxamer thermophoretic motion displays a remarkable double sign inversion, which is fully correlated with a peak in the thermal expansivity of the solution marking the progressive dehydration of the propylene oxide groups of P407 and their incorporation into the micellar core. This rather puzzling behaviour of the thermophoretic mobility and of the Soret coefficient in the P407 micellization region can tentatively be explained by properly taking into account the temperature-dependent balance between micellized and nonassociated poloxamer chains.
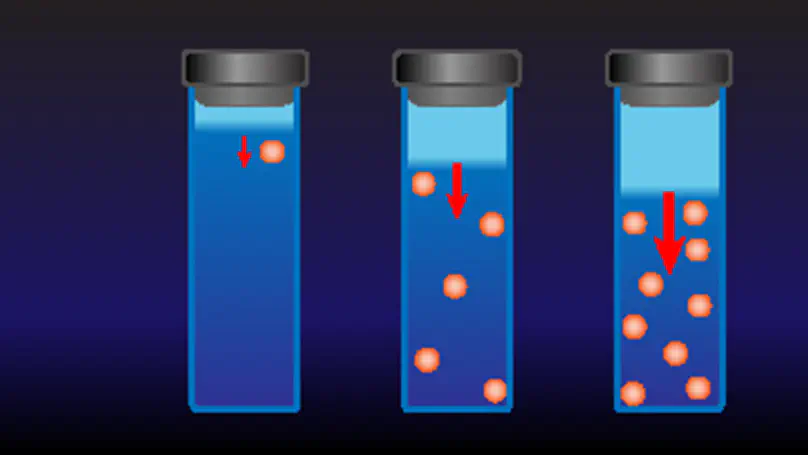
By experimenting on model colloids where depletion forces can be carefully tuned and quantified, we show that attractive interactions consistently “promote” particle settling, so much that the sedimentation velocity of a moderately concentrated dispersion can even exceed its single-particle value. At larger particle volume fraction $\phi$, however, hydrodynamic hindrance eventually takes over. Hence, $v(\phi)$ actually displays a nonmonotonic trend that may threaten the stability of the settling front to thermal perturbations. Finally, by discussing a representative case, we show that these results are relevant to the investigation of protein association effects by ultracentrifugation.
Recent Publications
Contact
- lattuada.enrico@gmail.com
- +39 346 2292 066
- Via E. Rainusso, 144, Modena, 41124